Variant DetailsVariant: nsv469675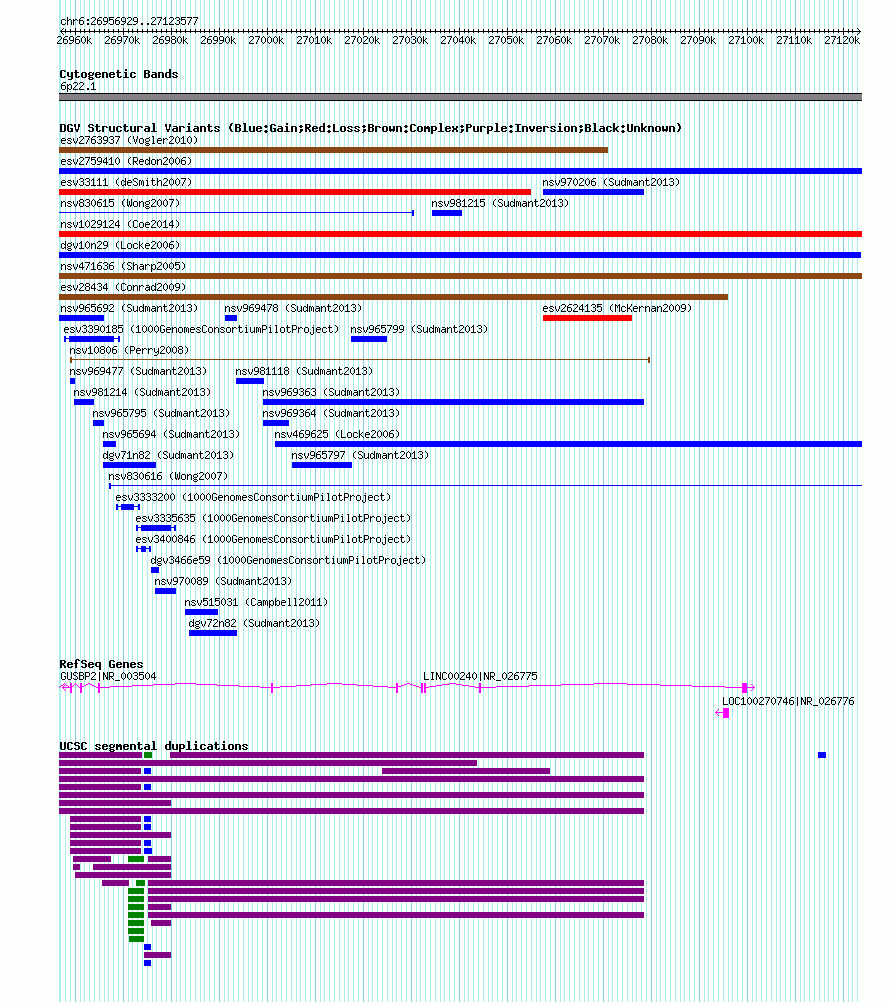 | Internal ID | 15187704 | | Landmark | | | Location Information | | | Cytoband | 6p22.1 | | Allele length | | Assembly | Allele length | | hg38 | 166649 | | hg19 | 166649 | | hg18 | 166649 | | hg16 | 166649 |
| | Variant Type | CNV gain | | Copy Number | | | Allele State | | | Allele Origin | | | Probe Count | | | Validation Flag | | | Merged Status | M | | Merged Variants | dgv10n29 | | Supporting Variants | nssv1676215, nssv1673324, nssv1676287, nssv1674490, nssv1675364, nssv1675208, nssv1673132, nssv1676404, nssv1672887, nssv1672141, nssv1672500, nssv1675659, nssv1675803, nssv1673743, nssv1676536, nssv1672836, nssv1674166, nssv1676498, nssv1673802, nssv1673258, nssv1674173, nssv1672607, nssv1672691, nssv1674626 | | Samples | | | Known Genes | GUSBP2, LINC00240, LOC100270746 | | Method | BAC aCGH | | Analysis | A locus was considered a CNV if the log ratio of fluorescence measurements for the individuals assayed exceeded twice the SD of the autosomal clones in both dye-swapped experiments. | | Platform | GPL4010 | | Comments | | | Reference | Locke_et_al_2006 | | Pubmed ID | 16826518 | | Accession Number(s) | nsv469675
| | Frequency | | Sample Size | 265 | | Observed Gain | 24 | | Observed Loss | 0 | | Observed Complex | 0 | | Frequency | n/a |
|
|
