Variant DetailsVariant: esv3590421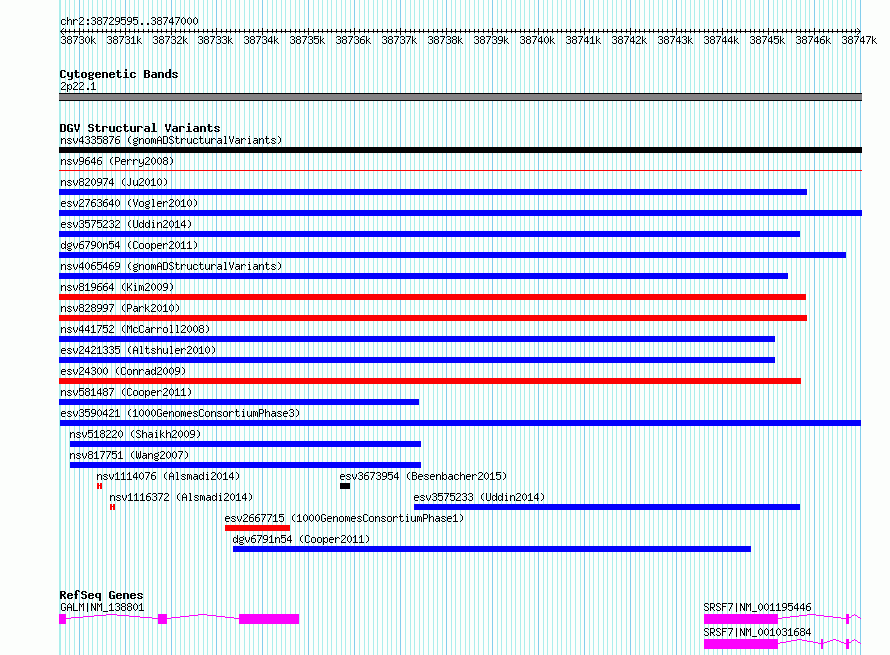 | Internal ID | 6630707 | | Landmark | | | Location Information | | | Cytoband | 2p22.1 | | Allele length | | Assembly | Allele length | | hg38 | 17406 | | hg19 | 17406 |
| | Variant Type | CNV gain | | Copy Number | | | Allele State | | | Allele Origin | | | Probe Count | | | Validation Flag | | | Merged Status | M | | Merged Variants | | | Supporting Variants | essv10468488, essv10468458, essv10468493, essv10468443, essv10468431, essv10468467, essv10468475, essv10468440, essv10468455, essv10468434, essv10468479, essv10468444, essv10468494, essv10468450, essv10468418, essv10468427, essv10468498, essv10468424, essv10468453, essv10468483, essv10468492, essv10468411, essv10468469, essv10468419, essv10468451, essv10468432, essv10468421, essv10468500, essv10468425, essv10468464, essv10468422, essv10468502, essv10468448, essv10468445, essv10468438, essv10468507, essv10468471, essv10468439, essv10468409, essv10468457, essv10468459, essv10468497, essv10468426, essv10468413, essv10468435, essv10468423, essv10468496, essv10468485, essv10468470, essv10468463, essv10468417, essv10468477, essv10468430, essv10468480, essv10468487, essv10468499, essv10468433, essv10468429, essv10468442, essv10468420, essv10468462, essv10468410, essv10468482, essv10468481, essv10468504, essv10468416, essv10468501, essv10468460, essv10468447, essv10468441, essv10468486, essv10468484, essv10468456, essv10468449, essv10468490, essv10468474, essv10468415, essv10468466, essv10468495, essv10468472, essv10468473, essv10468454, essv10468468, essv10468478, essv10468446, essv10468412, essv10468465, essv10468428, essv10468476, essv10468452, essv10468505, essv10468503, essv10468461, essv10468436, essv10468506, essv10468489, essv10468437, essv10468491, essv10468414 | | Samples | HG00096, NA19794, HG00102, HG01610, HG00384, NA10851, HG00351, HG01079, HG00100, HG04194, HG01586, HG03767, NA11931, NA19734, HG00257, HG00315, HG00306, HG00318, HG00244, HG01686, HG01531, HG00337, HG00327, HG00641, HG04100, NA12812, HG01070, HG01351, HG02485, HG00173, NA12348, HG00736, NA07347, NA12283, HG01510, NA20287, HG00311, NA19923, NA12005, HG03817, HG02315, HG00379, HG00335, HG00106, HG01405, HG02427, HG01771, HG01275, HG00178, NA20757, NA20587, HG01699, HG00260, HG01353, HG00313, HG00137, HG00149, HG00731, HG00380, HG00282, HG01271, NA20521, NA20126, NA20832, HG01092, HG03742, HG02604, HG01707, NA12249, NA12827, HG03854, HG01334, HG03672, NA12778, HG00336, HG02220, HG00357, HG01108, NA12763, NA06994, HG01432, HG00381, HG00342, HG00186, HG01631, HG00280, HG00131, HG02052, HG00252, HG00105, NA20503, NA11892, HG01775, HG01111, HG03890, NA07000, HG01097, HG01786, HG03886 | | Known Genes | GALM, SRSF7 | | Method | Sequencing | | Analysis | | | Platform | Multiple platforms | | Comments | | | Reference | 1000_Genomes_Consortium_Phase_3 | | Pubmed ID | 21293372 | | Accession Number(s) | esv3590421
| | Frequency | | Sample Size | 2504 | | Observed Gain | 99 | | Observed Loss | 0 | | Observed Complex | 0 | | Frequency | n/a |
|
|
